Mario Valerio Giuffrida, Peter Doerner, Sotirios A. Tsaftaris
The Plant Journal (2018)
Mario Valerio Giuffrida, Peter Doerner, Sotirios A. Tsaftaris (2018) “Pheno-Deep Counter: a unified and versatile deep learning architecture for leaf counting,” The Plant Journal.
Abstract
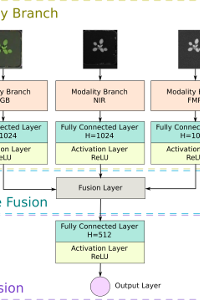
Direct observation of morphological plant traits is tedious and a bottleneck for high-throughput phenotyping. Hence, interest in image-based analysis is increasing, requiring software that can reliably extract plant traits, such as leaf count, preferably across a variety of species and growth conditions. However, current leaf counting methods do not work across species or conditions and therefore may broad utility. In this paper, we present Pheno-Deep Counter, a single deep network that can predict leaf count in 2D plant images of different species with rosette-shaped appearance. We demonstrate that our architecture can count leaves from multi-modal 2D images, such as RGB, fluorescence, and near-infrared. Our network design is flexible, allowing for inputs to be added or removed to accommodate new modalities. Furthermore, our architecture can be used as is without requiring dataset-specific customization of the internal structure of the network, opening its use to new scenarios. Pheno-Deep Counter is able to produce accurate predictions in many plant species and, once trained, can count leaves in a few seconds. Through our universal and open source approach to deep counting, we aim to broaden utilization of machine learning based approaches to leaf counting. Our implementation can be downloaded at https://bitbucket.org/tuttoweb/pheno-deep-counter.