Andrei Dobrescu, Mario Valerio Giuffrida, Sotirios A. Tsaftaris
Frontiers in Plant Science (2020)
Andrei Dobrescu, Mario Valerio Giuffrida, Sotirios A. Tsaftaris (2020) “Doing More With Less: A Multi-Task Deep Learning Approach in Plant Phenotyping,” Frontiers in Plant Science.
Abstract
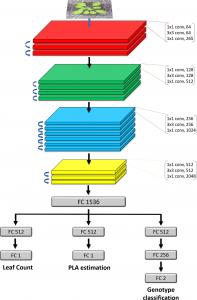
Image-based plant phenotyping has been steadily growing and this has steeply increased the need for more efficient image analysis techniques capable of evaluating multiple plant traits. Deep learning has shown its potential in a multitude of visual tasks in plant phenotyping, such as segmentation and counting. Here, we show how different phenotyping traits can be extracted simultaneously from plant images, using multitask learning (MTL). MTL leverages information contained in the training images of related tasks to improve overall generalization and learns models with fewer labels. We present a multitask deep learning framework for plant phenotyping, able to infer three traits simultaneously: (i) leaf count, (ii) projected leaf area (PLA), and (iii) genotype classification. We adopted a modified pretrained ResNet50 as a feature extractor, trained end-to-end to predict multiple traits. We also leverage MTL to show that through learning from more easily obtainable annotations (such as PLA and genotype) we can predict a better leaf count (harder to obtain annotation). We evaluate our findings on several publicly available datasets of top-view images of Arabidopsis thaliana. Experimental results show that the proposed MTL method improves the leaf count mean squared error (MSE) by more than 40%, compared to a single task network on the same dataset. We also show that our MTL framework can be trained with up to 75% fewer leaf count annotations without significantly impacting performance, whereas a single task model shows a steady decline when fewer annotations are available. Code available at https://github.com/andobrescu/Multi_task_plant_phenotyping.